User Guide
Running rMAPS
rMAPS consists of two tools. Motif Map performs motif enrichment analysis for RNA-binding proteins. CLIP Map examines CLIP-seq peak data to generate RNA-map of the protein used in the CLIP-seq experiment.
Running Motif Map
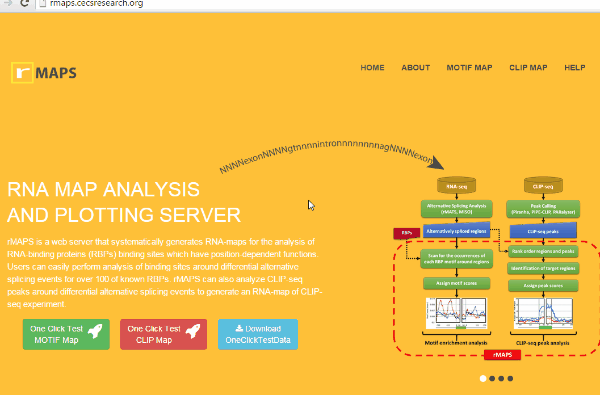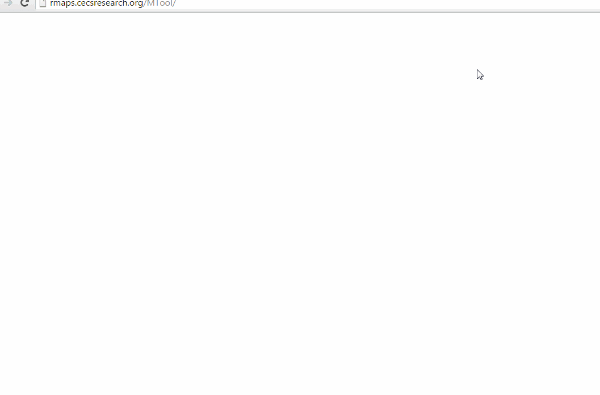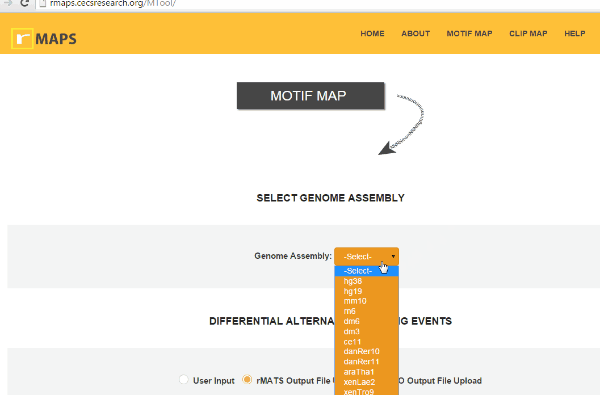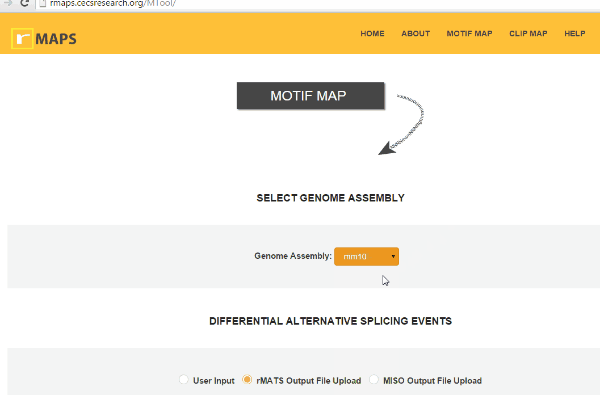
 Select Genome Assembly
Select Genome AssemblyIt is a simple step. You can simply select Genome assembly used in the RNA-seq experiment. rMAPS currently supports human (hg38, hg19), mouse (mm10), fly (dm6, dm3), rat (rn6), C. elegans (ce11), zebrafish (danRer10, danRer11), Arabidopsis thaliana (araTha1), frog (xenLae2, xenTro9), and rice (oSa7) are available.
 Upload Your Differential Alternative Splicing Output Data
Upload Your Differential Alternative Splicing Output DatarMAPS supports rMATS outputs, miso outputs, and user-provided coordinates for differential alternative splicing exons and control exons. rMAPS can analyze all 5 major types of alternative splicing events as follows:
Skipped Exon (SE)
Mutually eXclusive Exon (MXE)
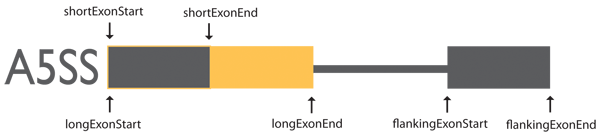
Alternative to 5′ Splice Site (A5SS)
Alternative to 3′ Splice Site (A3SS)
Retained Intron (RI)
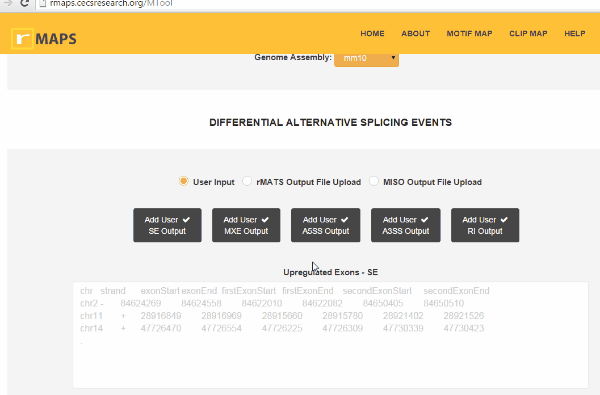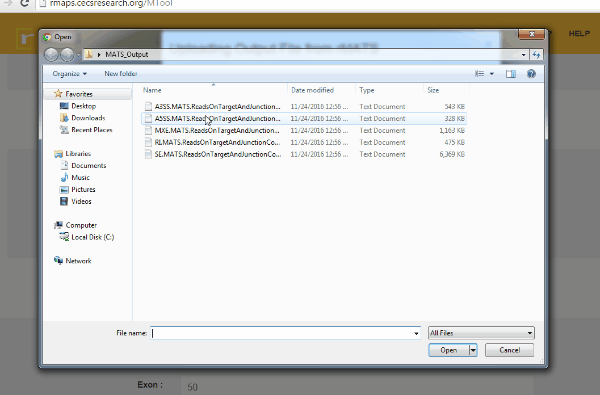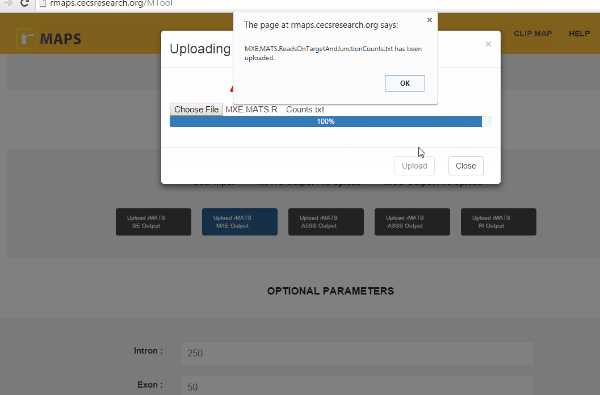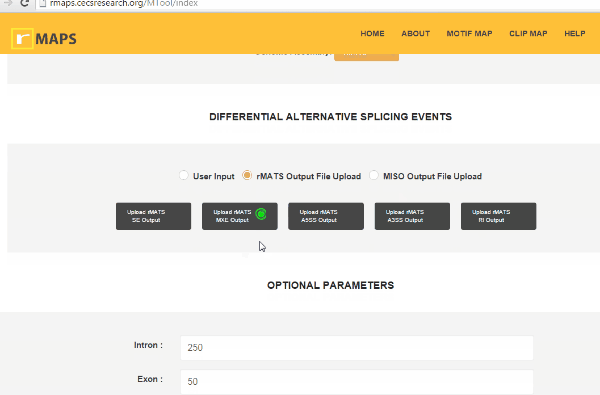
- Upload rMATS output (simple and recommended).
- Run rMATS program (computational tool to detect differential alternative splicing events).
- Save output files from the rMATS run.
- Click on “upload” then select files for alternative splicing events that you are interested in:
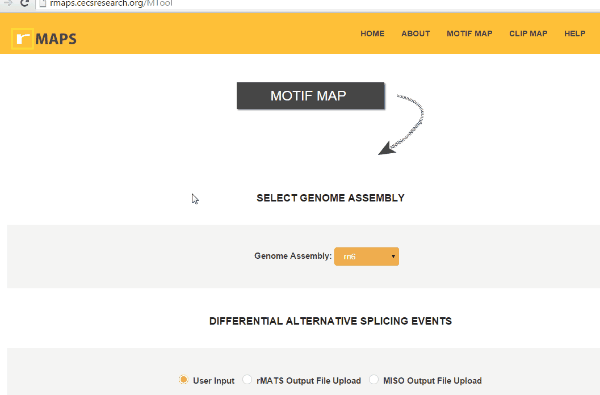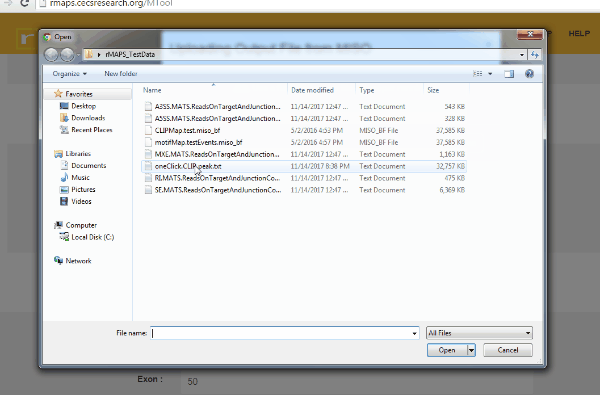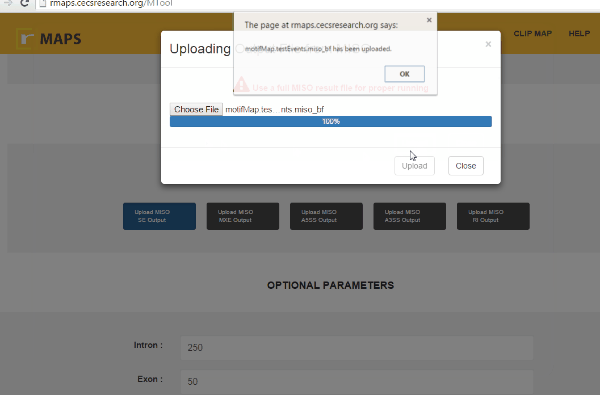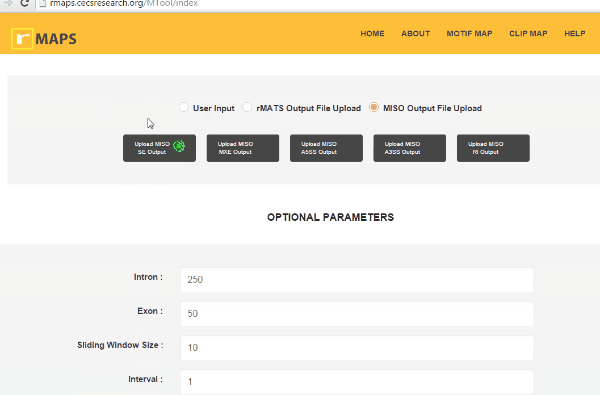
- Upload MISO output.
- Run MISO program (computational tool to detect differential alternative splicing events).
- Save output files from the MISO run.
- Click on “upload” then select files for alternative splicing events that you are interested in:
- Upload coordinates manually.
- Provide coordinates of upregulated exons, downregulated exons, background exons for alternative splicing events that you are interested in:

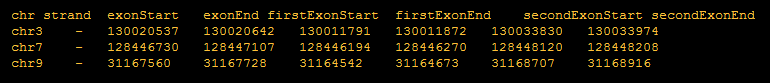
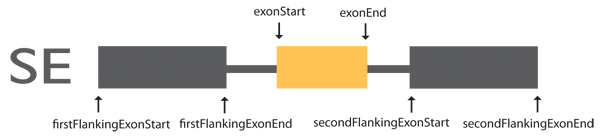
Exon coordinates are from three exons involved in skipping events.

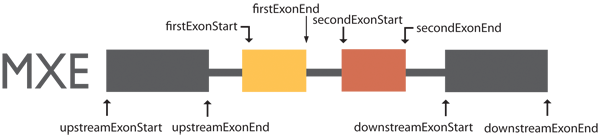
Exon coordinates are from four exons involved in mutually exclusive events.
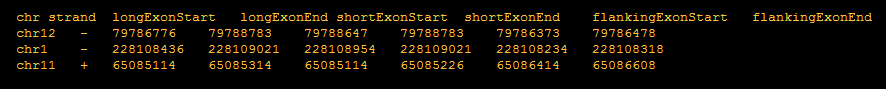
Exon coordinates are from three exons involved in alternative to 5′ splicing events.

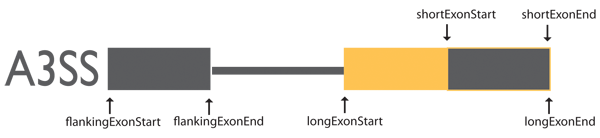
Exon coordinates are from three exons involved in alternative to 3′ splicing events.

Exon coordinates are from three exons involved in retained intron events.

 Enter “Optional Parameters”
Enter “Optional Parameters”- Intron (default 250): Length of the intronic region to be examined and plotted.
- Exon (default 50): Length of the exonic region to be examined and plotted.
- Sliding Window Size (default 50): Size of the sliding window to be used to count the motif occurrences.
- Interval (default 1): Step size of windows sliding.
- Additional Motif (no default value): User input of Motif
-
rMAPS takes optional motifs from users with flexibility provided by the regular expressions.
Here are some useful regular expressions with examples.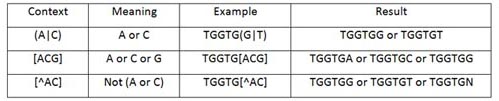
Multiple regular expressions can be used at the same time.
For example, [TG]TGG[GC]T would look for 4 patterns
(i.e., TTGGGT or TTGGCT or GTGGGT or GTGGCT) - Email (no default value): An option to send a link of the result via email
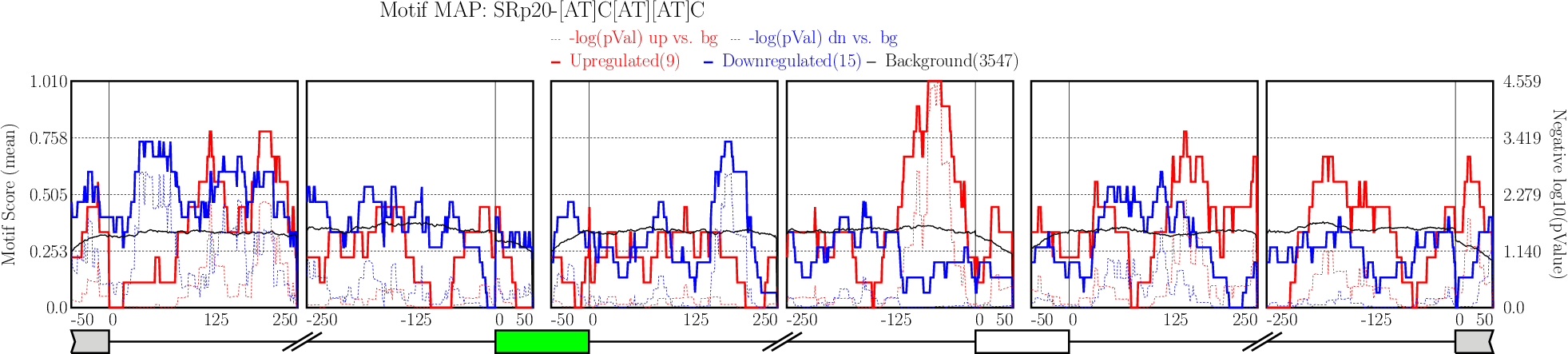
 Click on “Run” button to get motif enrichment analysis plots.
Click on “Run” button to get motif enrichment analysis plots.It typically takes under 4 minutes for all 5 alternative splicing types together.
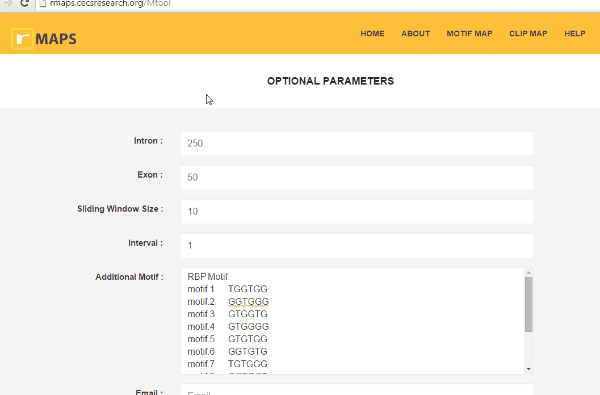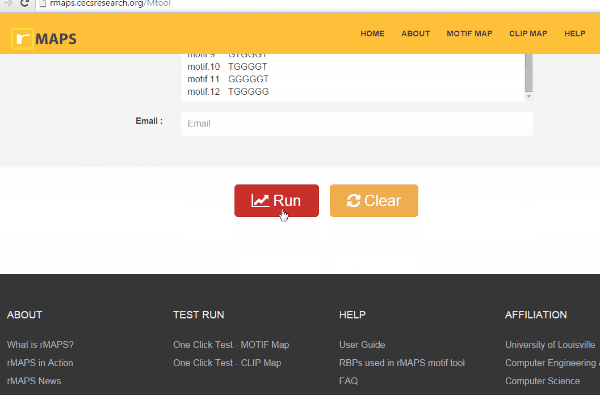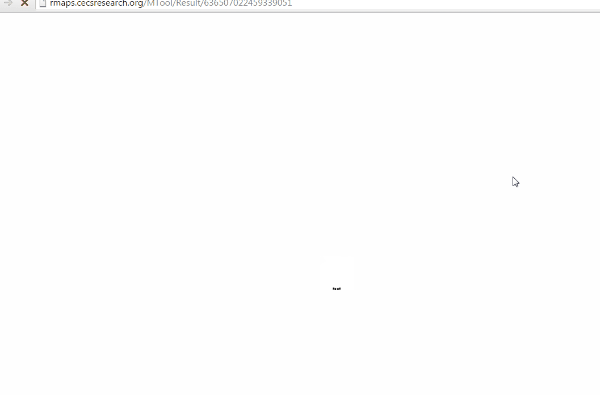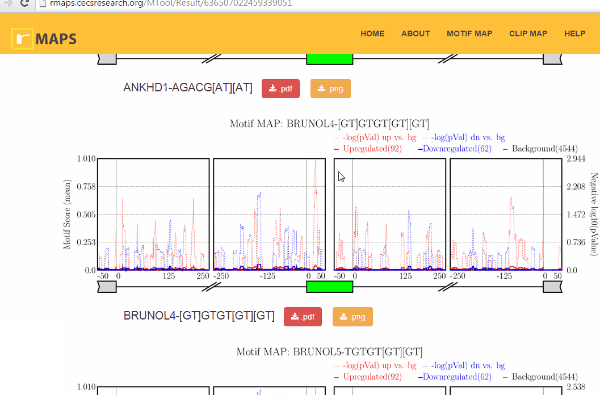
 Examine output plots and folder.
Examine output plots and folder.- rMAPS gives a link to output folder which will be accessible for 48 hours.
- rMAPS also provides links to each plot in both PDF format and PNG format.

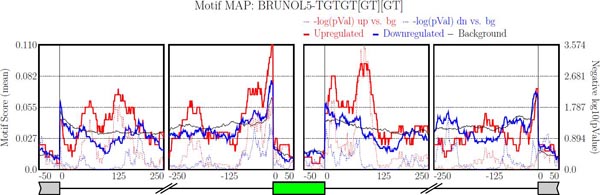
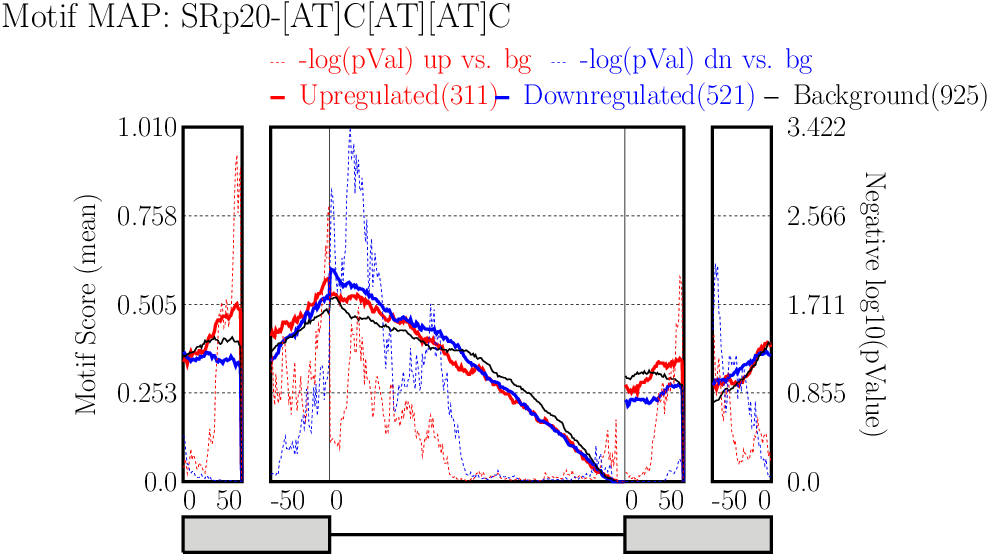
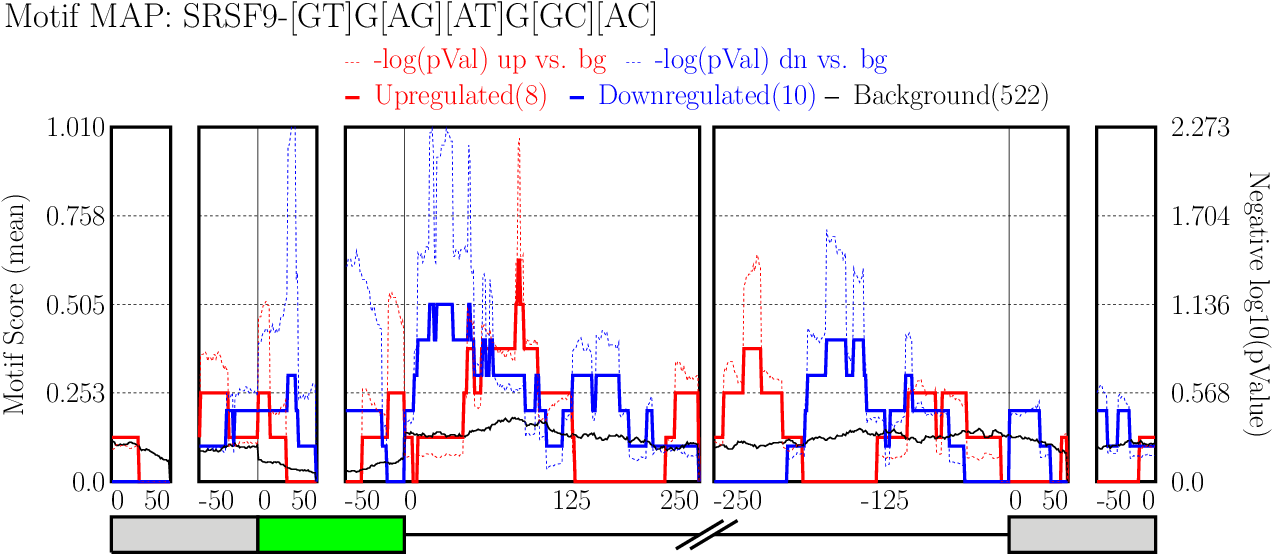
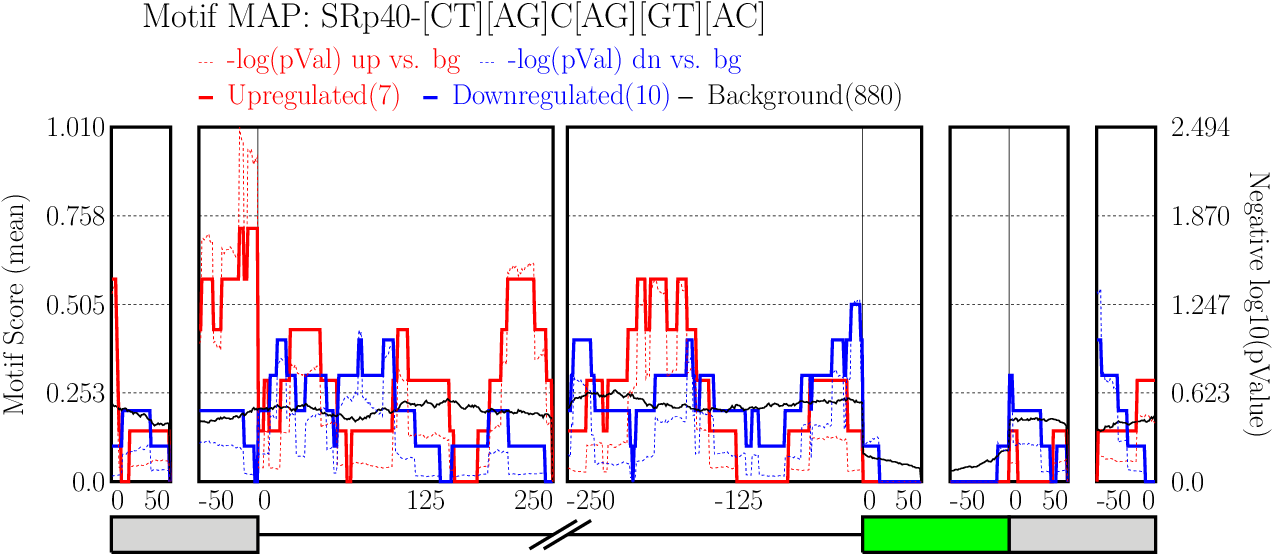
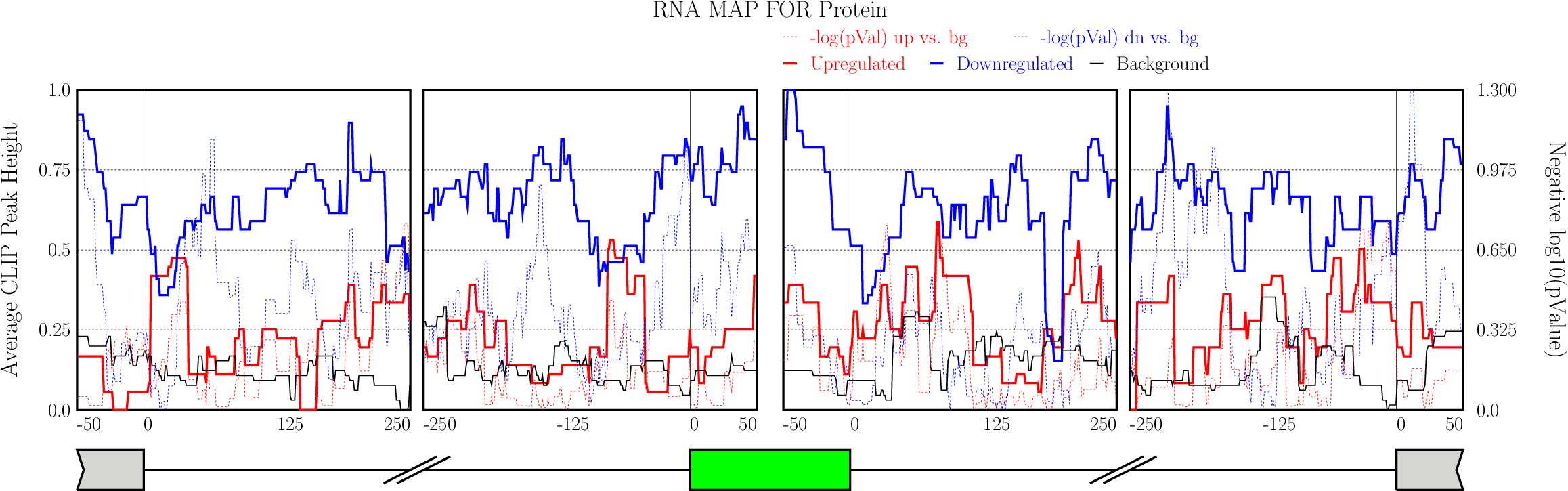
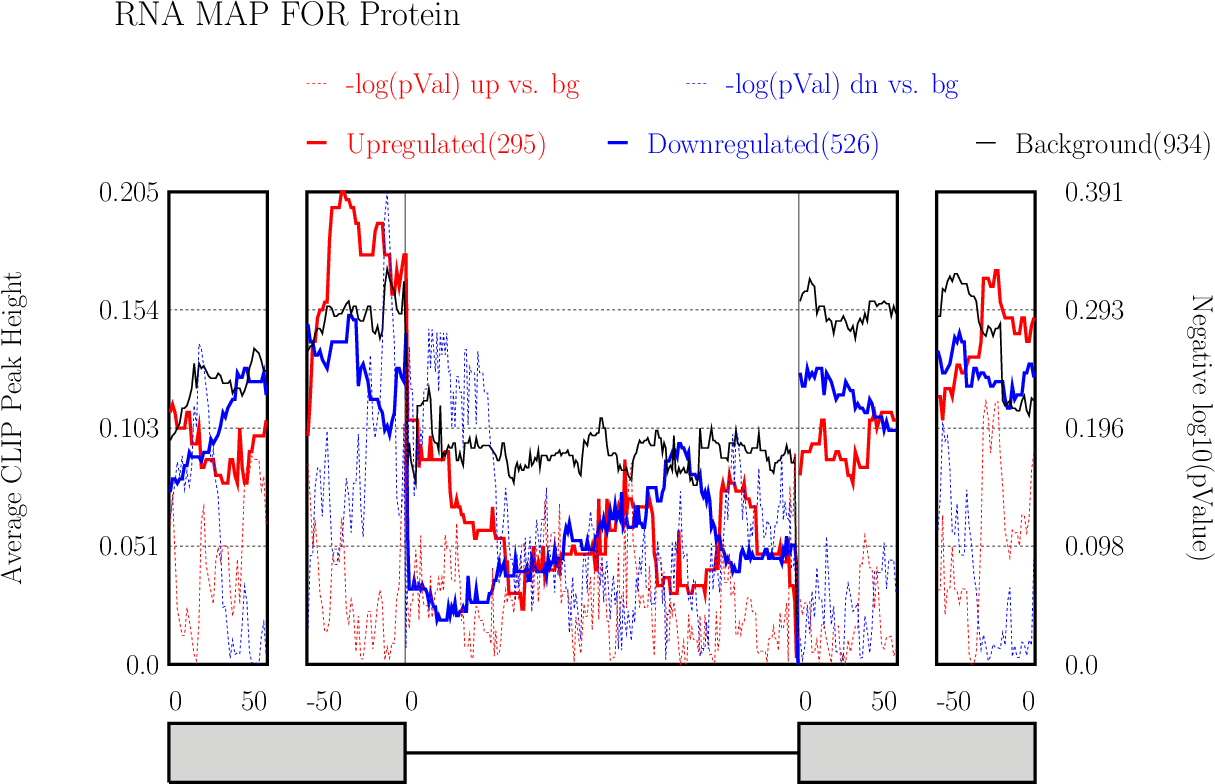
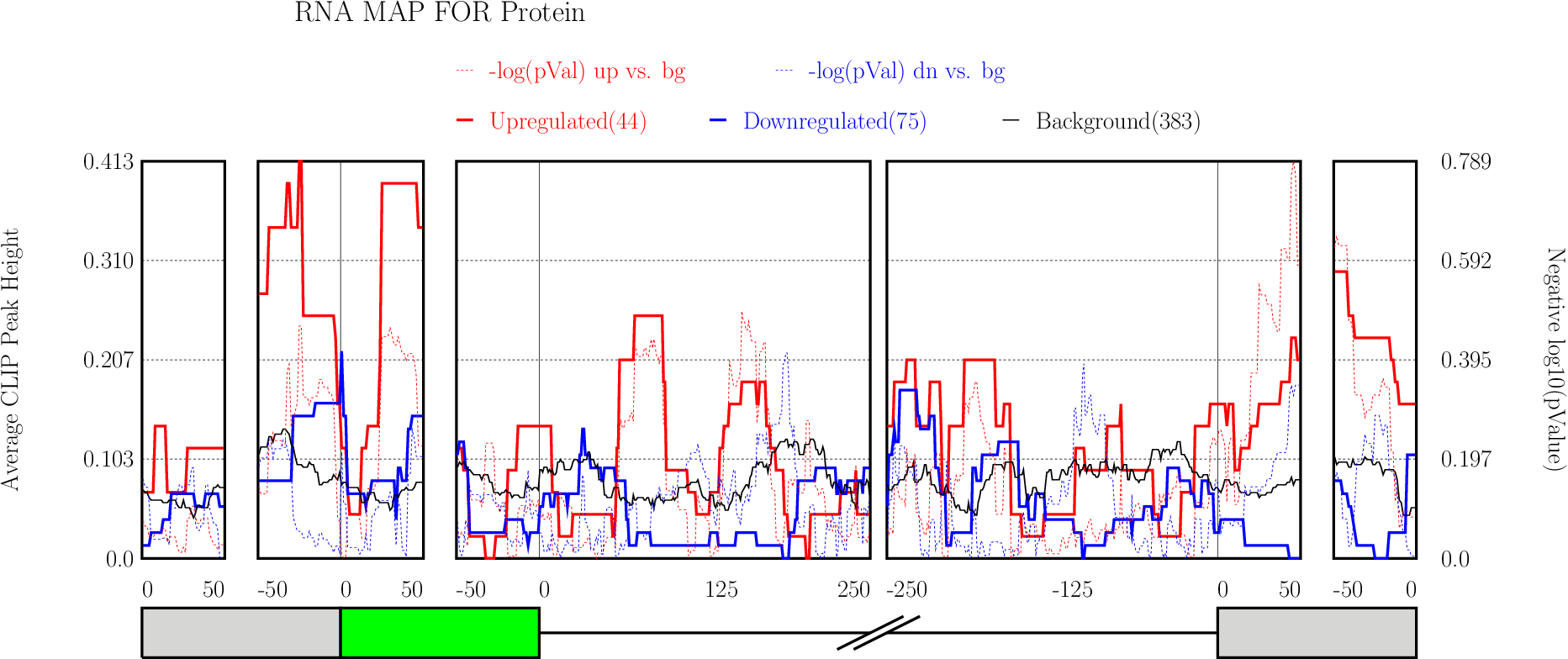
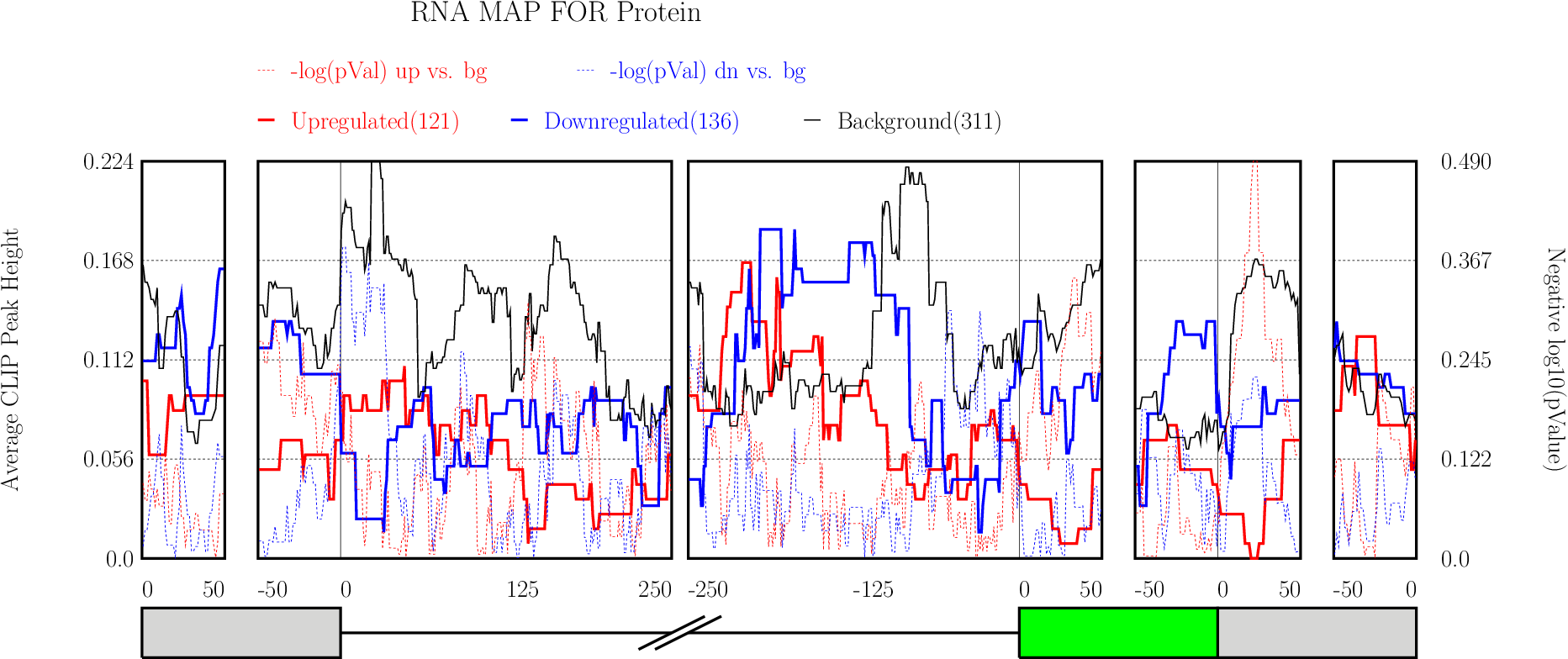
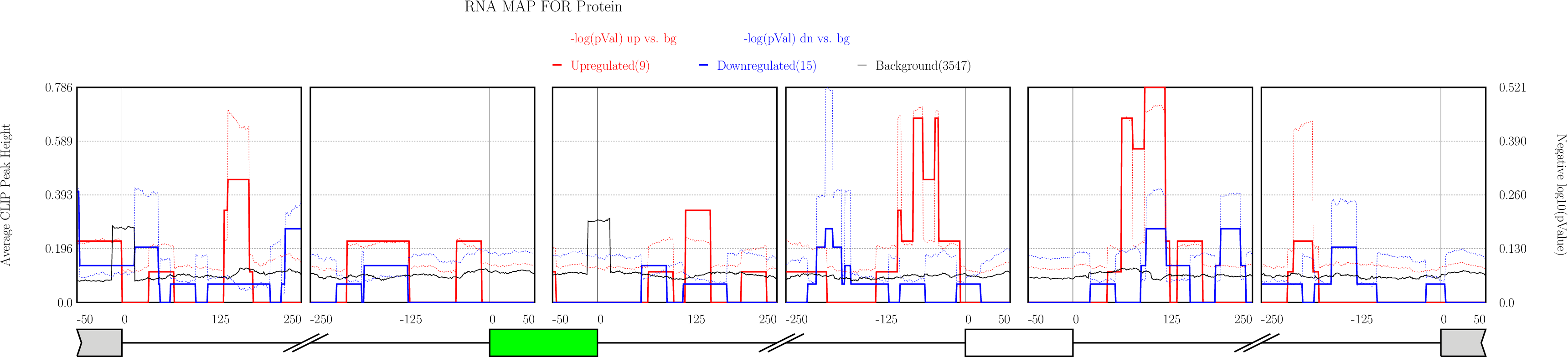
Running CLIP Map
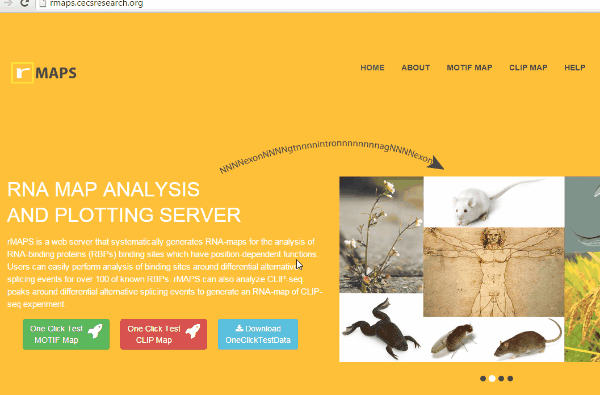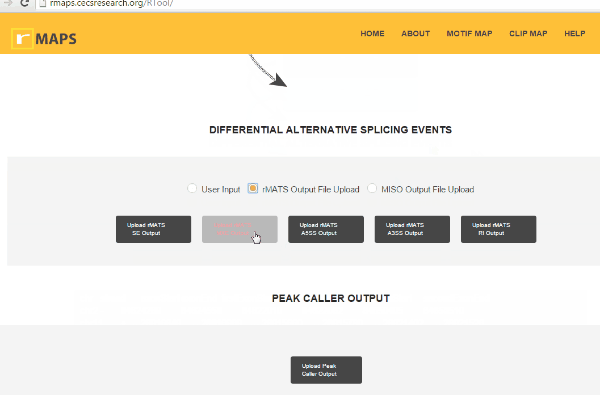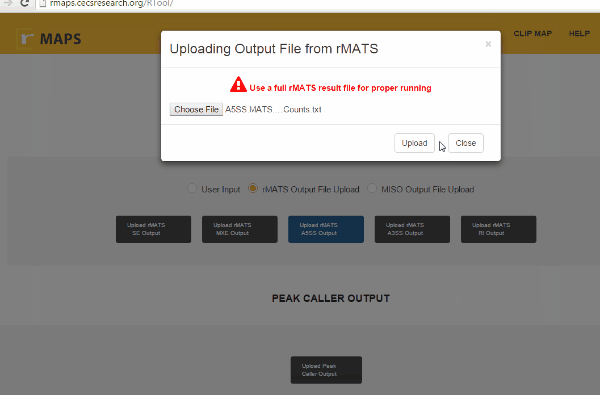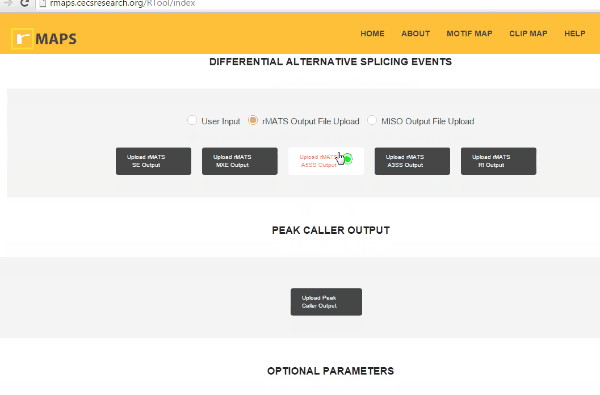
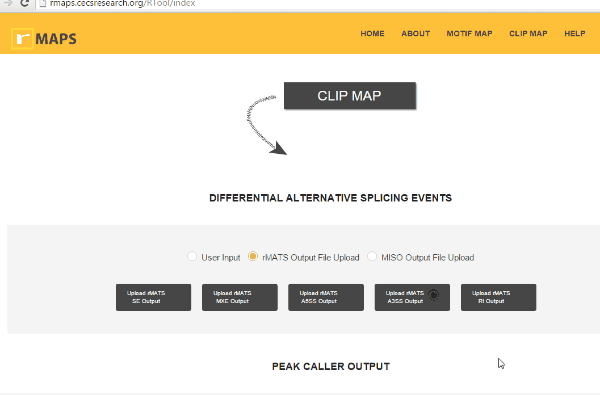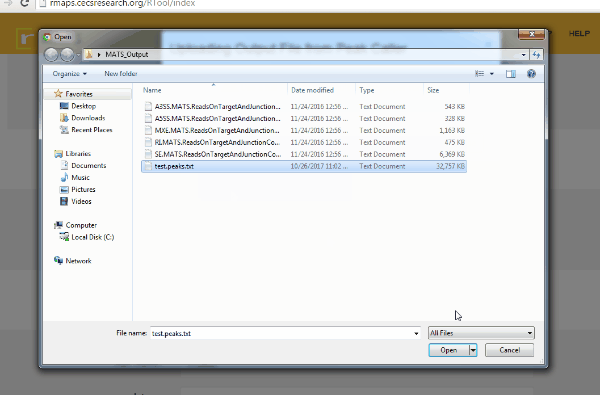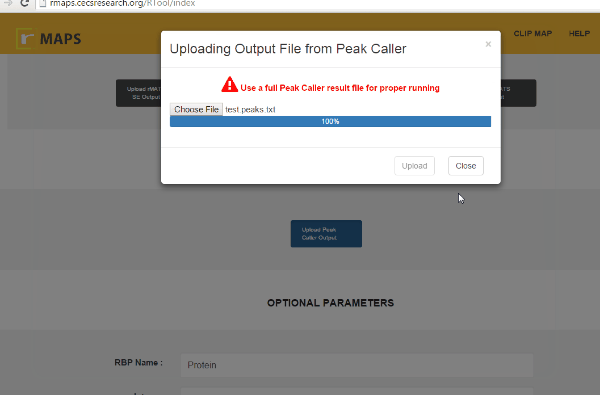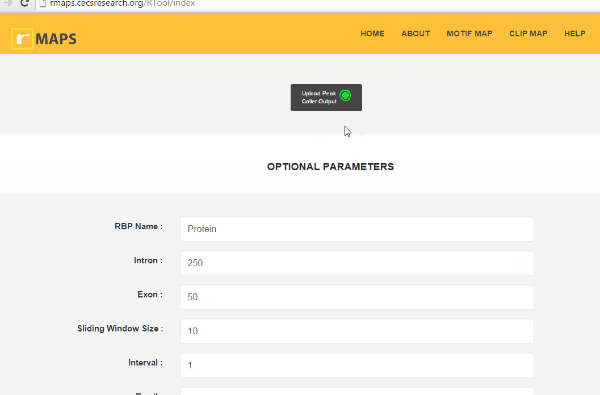
 Upload Your Differential Alternative Splicing Output Data
Upload Your Differential Alternative Splicing Output Data- Upload rMATS output (simple and recommended).
- Run rMATS program (computational tool to detect differential alternative splicing events).
- Save output files from the rMATS run.
- Click on “upload” then select files for alternative splicing events that you are interested in:
 Enter “Optional Parameters”.
Enter “Optional Parameters”.- RBP Name (default Protein): Enter the name of the protein used in CLIP-seq experiment.
- Intron (default 250): Length of the intronic region to be examined and plotted.
- Exon (default 50): Length of the exonic region to be examined and plotted.
- Sliding Window Size (default 50): Size of the sliding window to be used to count the motif occurrences.
- Interval (default 1): Step size of windows sliding.
- Email (no default value): An option to send a link of the output result via email
 Click on “Run” button get RNA-map analysis plots.
Click on “Run” button get RNA-map analysis plots.It typically takes under 1 minute for all 5 alternative splicing types together.
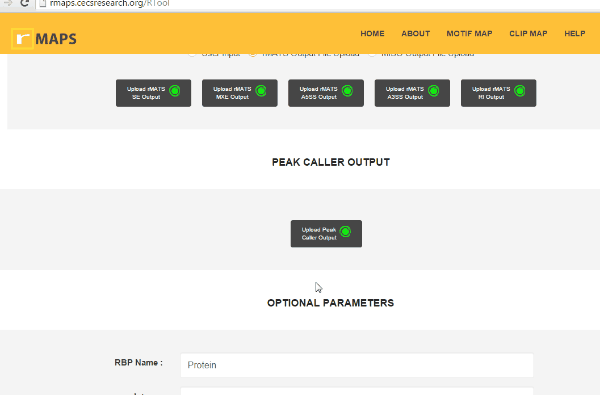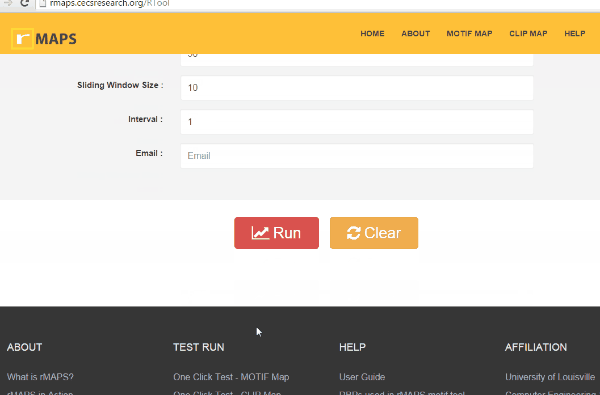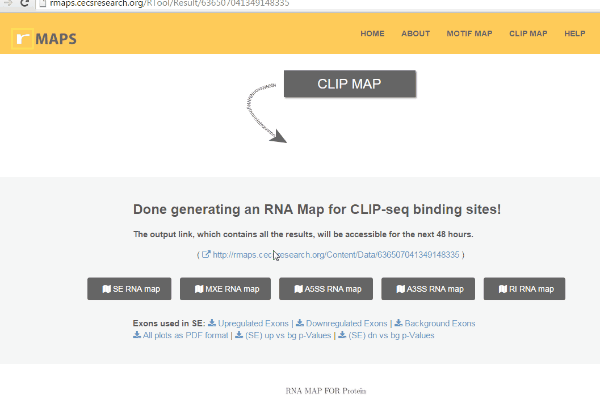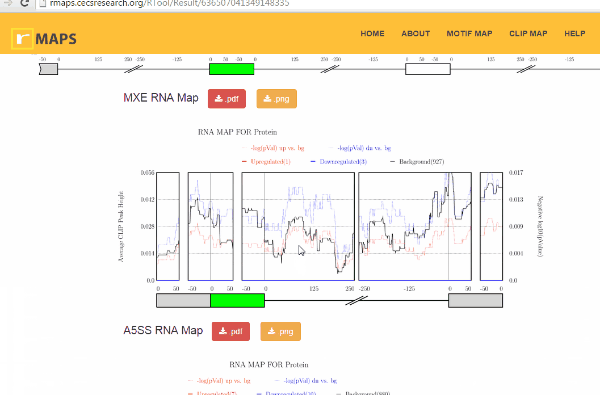
 Examine output plots and folder.
Examine output plots and folder.- rMAPS gives a link to output folder which will be accessible for 48 hours.
- rMAPS also provides links to each plot in both PDF format and PNG format.